Introduction
Although {dplyover} is an extension of {dplyr}, it doesn’t make use of (read: copy) {dplyr} internals. This made it relatively easy to develop the package without (i) copying tons of {dplyr} code, (ii) having to figure out which dplyr-functions use the copied internals and (iii) finally overwritting these functions (like mutate and other one-table verbs), which would eventually lead to conflicts with other add-on packages, like for example {tidylog}.
However, the downside is that not relying on {dplyr} internals has some negative effects in terms of performance and compability.
In a nutshell this means:
- The over-across function family in {dplyover} is slower than the original
dplyr::across. Up until {dplyr} 1.0.3 the overhead was not too big, butdplyr::acrossgot much faster with {dplyr} 1.0.4 which why the gap has widend a lot. - Although {dplyover} is designed to work in {dplyr}, some features and edge cases will not work correctly.
The good news is that even without relying on {dplyr} internals most of the original functionality can be replicated, and although being less performant, the current setup is optimized and falls not too far behind in terms of speed - at least when compared to the 1.0.3 <= version of dplyr::across.
Regarding compability, I have spent quite some time testing the package and I was able to replicate most of the tests for dplyr::across successfully.
Below both issues, compability and performance, are addressed in more detail.
Before we begin, here is our setup:
Compability
At the moment three unintended compability issues are worth mentioning:
Only partial support of mutate()’s .keep argument
Since {dplyr} 1.0 mutate has gained a .keep argument, which allows us to control which columns are retained in the output. This works well with genuine {dplyr} functions like across:
diamonds %>% mutate(across(z, ~ .x + y),
.keep = "used")
#> # A tibble: 53,940 x 2
#> y z
#> <dbl> <dbl>
#> 1 3.98 6.41
#> 2 3.84 6.15
#> 3 4.07 6.38
#> 4 4.23 6.86
#> # ... with 53,936 more rows.keep not only recognizes that across is used on colum ‘z’, it also registers that we used column ‘y’ in across’s .fns argument. Unfortunately, .keep is only partially supported within {dplyover}. It works with columns that are used inside the .fns argument.
diamonds %>% mutate(over(1, ~ y + 1),
.keep = "used")
#> # A tibble: 53,940 x 2
#> y `1`
#> <dbl> <dbl>
#> 1 3.98 4.98
#> 2 3.84 4.84
#> 3 4.07 5.07
#> 4 4.23 5.23
#> # ... with 53,936 more rowsBut it does not work with columns provide in the .xcols or .ycols argument in crossover or across2.
diamonds %>% mutate(crossover(y, 1, ~ .x + z),
.keep = "unused")
#> Warning: `crossover()` does not support the `.keep` argument in
#> `dplyr::mutate()` when set to 'used' or 'unused'.
#> # A tibble: 53,940 x 1
#> y_1
#> <dbl>
#> 1 6.41
#> 2 6.15
#> 3 6.38
#> 4 6.86
#> # ... with 53,936 more rowsFor this reason, crossover, across2, and across2x issue a warning when mutate’s .keep argument is specified.
Differences in the glue syntax
dplyr::across() allows the use of local variables in addition to {col} and {fn} in the glue syntax of its .names argument. This is currently not supported by the over-across functions in {dplyover}. To somewhat leviate this issue the over-across functions allow character vectors to be supplied to the .names argument (similar to poorman::across).
Example:
We can do this with dplyr::across:
prefix <- "lag1"
tibble(a = 1:25) %>%
mutate(across(everything(), lag, 1, .names = "{.col}_{prefix}"))
#> # A tibble: 25 x 2
#> a a_lag1
#> <int> <int>
#> 1 1 NA
#> 2 2 1
#> 3 3 2
#> 4 4 3
#> # ... with 21 more rowsIn {dplyover} we would need to construct the names outside of the function call and supply this vector to .names:
col_nms <- "a_lag1"
tibble(a = 1:25) %>%
mutate(over(1, ~ lag(a, .x), .names = col_nms))
#> # A tibble: 25 x 2
#> a a_lag1
#> <int> <int>
#> 1 1 NA
#> 2 2 1
#> 3 3 2
#> 4 4 3
#> # ... with 21 more rowsApart from supplying an external character vector to .names the over-across functions have a special set of glue specifications that give more control over how vectors are named. This should minimize the need to construct the names externally and supply them as a vector. Please refer to the documentation of each function to learn more about which special glue specifications are supported.
Context dependent expressions: cur_column()
When over-across functions are called inside dplyr::mutate or dplyr::summarise {dplyr}’s context dependent expressions (see ?dplyr::context) can be used inside the function call. An exception is dplyr::cur_column() which works only inside dplyr::across and is neither supported in across2 nor crossover.
It is likely that there are more edge cases in which {dplyover}’s over-across functions are behaving differently (in an unintended way) from its relative dplyr::across. Feel free to contact me or just open an issue on GitHub.
Performance
{dplyover}’s performance issues are discussed in two steps. First, we compare major {dplyover} functions with dplyr::across and look at the performance of the internal setup. Since this is a rather theoretical comparison, we then examine an actual operation using {dplyover}’s crossover and compare its performance to existing workarounds.
Internal setup
To compare the performance of dplyr::across with the over-across functions from {dplyover} we take the diamonds data set from the {ggplot2} package. Since we are only interested in comparing the efficiency of the internal setup, we just loop over a couple of columns / elements and apply a function returning 1 to make sure that no computation is involved. Finally, we use the .names argument to name the output columns.
diamonds %>%
summarise(across(c(x,y,z),
~ 1,
.names = "{col}_new"))
diamonds %>%
summarise(over(c("q","v","w"),
~ 1,
.names = "{x}_new"))#> # A tibble: 2 x 3
#> expression median mem_alloc
#> <bch:expr> <bch:tm> <bch:byt>
#> 1 over 2.69ms 428KB
#> 2 across 4.44ms 734KBIf we compare the performance of both operations, we can see that over is slightly faster than dplyr::across. This changes, however, when we consider grouped data. To demonstrate this, we take the diamonds data set and create four versions: an ungrouped version, one with 10, 50 and one with 100 groups. Then we compare the same operations again. Now, we also add crossover and across2 to the benchmark.
Below is a code snippet for the 100 groups case:
diamonds_100grp <- diamonds %>%
group_by(grp_id = row_number() %% 100)
diamonds_100grp %>%
summarise(across(c(x,y,z),
~ 1,
.names = "{col}_new"))
diamonds_100grp %>%
summarise(over(c("q","v","w"),
~ 1,
.names = "{x}_new"))
diamonds_100grp %>%
summarise(crossover(c(x, y, z),
c(1:3),
~ 1,
.names = "{xcol}_{y}"))
diamonds_100grp %>%
summarise(across2(c(x, y, z),
c(x, y, z),
~ 1,
.names = "{xcol}_{ycol}"))The plot below shows the median time in miliseconds that each operation takes by increasing group size. Obviously, dplyr::across is by far the fastest (especially after dplyr v.1.0.4). over is somewhat slower and the least performant operations are crossover and across2. The latter two need to access the underlying data and without using {dplyr}’s data mask there seems to be no good option to do that.
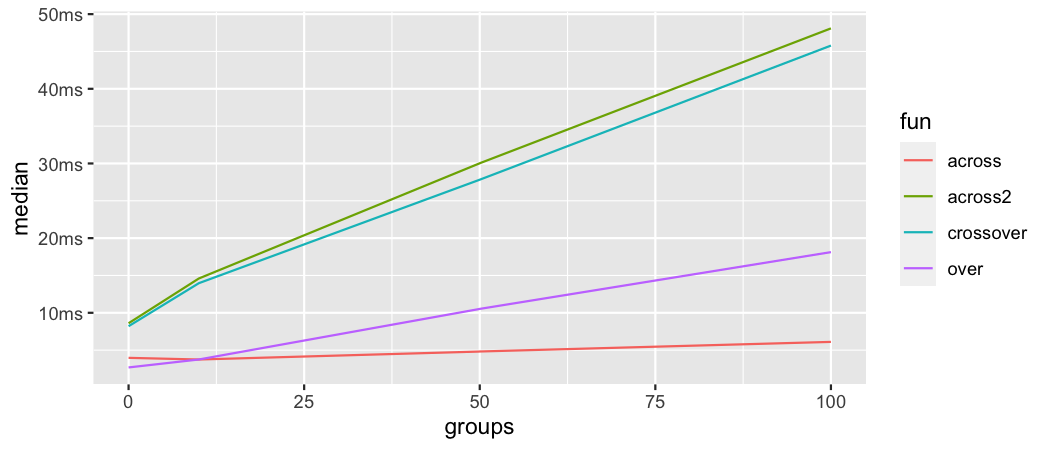
Benchmark of major dplyover functions
The performance of an actual operation
After getting a feeling for the general performance of the different functions in the over-across family we next have a look at an actual operation and how it performs compared to available workarounds.
Below we come back to the second example from the vignette “Why dplyover?”. We want to create several lagged variables for a set of columns.
We compare crossover with two alternative approaches:
- A call to
purrr:map_dfcnested insidedplyr::acrossand - A call to
reduce2in combination with a custom function.
# crossover
diamonds %>%
mutate(crossover(c(x,y,z),
1:5,
list(lag = ~ lag(.x, .y)),
.names = "{xcol}_{fn}{y}"))
# across and map_dfc
diamonds %>%
mutate(across(c(x,y,z),
~ map_dfc(set_names(1:5, paste0("lag", 1:5)),
function(y) lag(.x, y))
)) %>%
do.call(data.frame, .)
# custom function with reduce
create_lags2 <- function(df, .x, .y) {
mutate(df, "{.x}_lag{.y}" := lag(!! sym(.x), .y))
}
diamonds %>%
reduce2(rep(c("x", "y", "z"), 5),
rep(1:5,3),
create_lags2,
.init = .)When used on ungrouped data map_dfc nested in across is the most performant approach, while using a custom function with reduce is the least performant. crossover is not too far off in terms of speed compared to the map_dfc approach.
#> # A tibble: 3 x 3
#> expression median mem_alloc
#> <bch:expr> <bch:tm> <bch:byt>
#> 1 crossover 18.6ms 16MB
#> 2 across_map_dfc 15.2ms 15.5MB
#> 3 reduce_cst_fct 51.3ms 15.6MBThis picture gets a little bit more complex when we compare each approach across different group sizes. In the lower range (up until about 15 groups) map_dfc nested in across is the fastest approach, with crossover being not much slower. With increasing group size crossover starts to outperform map_dfc up until around 60 groups where reduce finally takes over and delivers the fastest performance. This benchmark stopped at 100 groups, but extrapolating from the existing data, we see that reduce is the most scaleable approach (although having the highest setup costs). If speed is a pressing concern, then the use of repetitive code patterns in a regular dplyr::across call is a valid option that is still faster than any of the approaches shown here.
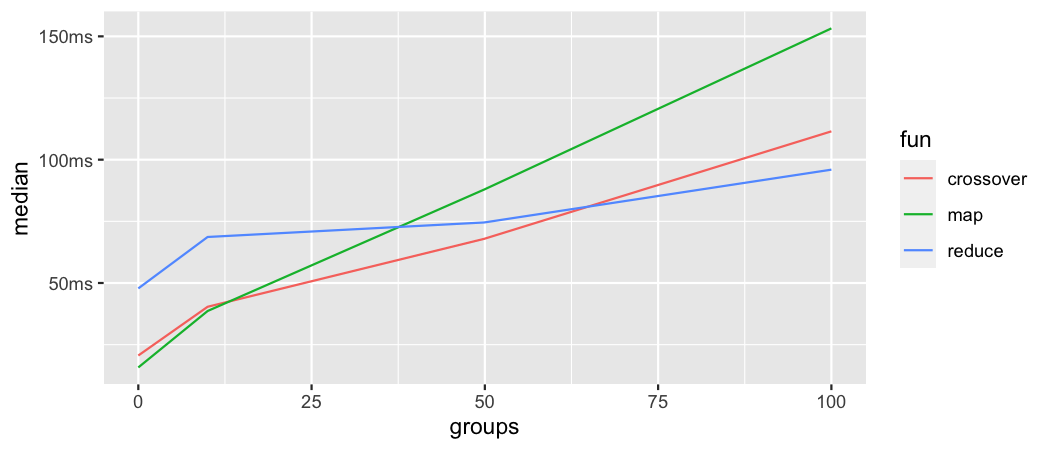
Benchmark: lagging many variables
Conclusion
Above we shed some light on {dplyover} performance when used on grouped data as well as its compability with {dplyr}. Regarding the latter, we only saw minor issues:
-
dplyr::mutate’s.keepargument can be easily replaced wih a call todplyr::select -
cur_column()might not make as much sense incrossoverandacross2as it does indplyr::acrossand - as an alternative to the use of local variables within the
.namesargument, all functions of the over-across family accept a character vector which can be used to construct the variables names.
Regarding the performance of the over-across functions when applied to grouped data we saw that they were far less performant than dplyr::across. However, when looking at an actual use case, we saw that the timings were quite reasonable compared to other programmatic alternatives.
Nevertheless, both issues, performance and compability, will be improved in future versions of {dplyover}.